Microbiotica accesses high quality large-scale clinical datasets in order to identify microbiome-based biomarkers of drug response, drug side-effects or disease progression. The company has accessed the clinical data from academic/clinical collaborations (University of Adelaide, Cambridge University Hospitals/Cancer Research UK) or from pharma partners (Genentech/Roche).
The Microbiotica platform, combining leading metagenomics, mass culturing, and machine learning, identifies gut microbial signatures linked to different patient outcomes. Bacterial signatures stratify patients for personalised drug treatment and also identify candidate bacteria at therapeutics or for target discovery.
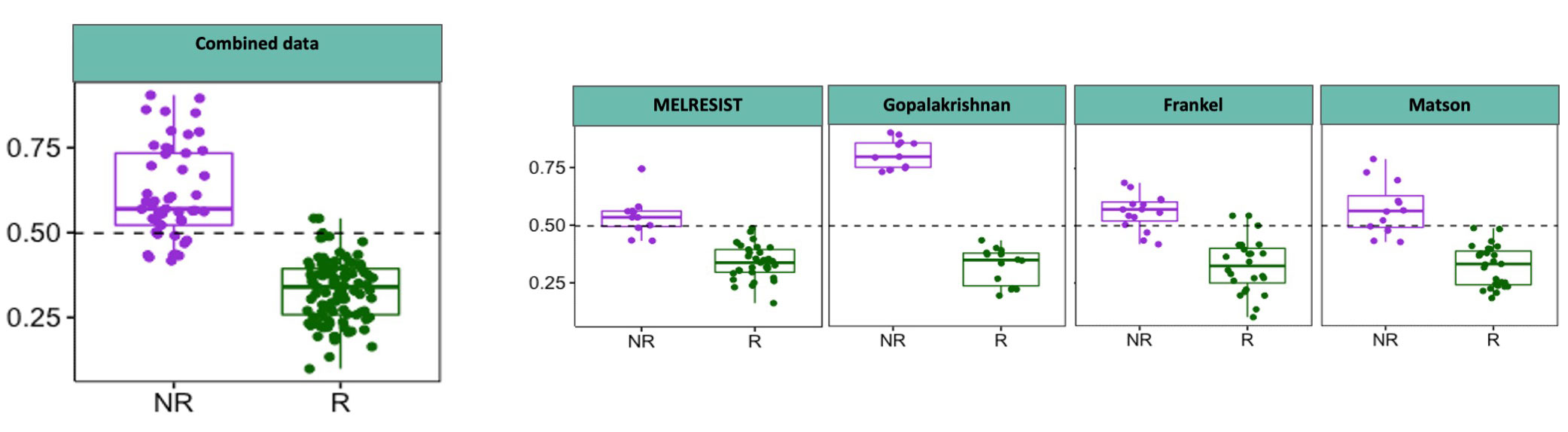
In an example using MELRESIST study data from our collaboration with Cambridge University Hospitals and Cancer Research UK, the company has identified a gut bacterial signature 91 % predictive of cancer immunotherapy response in melanoma. This signature is also predictive within three other melanoma studies and is the first microbiome signature predictive across multiple studies. It therefore represents a potential biomarker for therapeutic stratification and the source of a Live Bacterial Therapeutic.
Microbiotica Identifies First Single Response Signature Across Melanoma Immunotherapy Studies
- MELRESIST plus Microbiotica platform key to identifying common signal across all of 4 melanoma datasets that predicts response with 91% accuracy
- Precision in read classification
- Proprietary bioinformatics pipeline to allow analysis across different studies
- Machine learning models to identify signature
- First robust microbiome signature identified that predicts whether patients will respond to immunotherapy across multiple studies
- Confirming that the microbiome is a key driver of immune checkpoint inhibitor response
- Signature primarily driven by bacteria elevated in responders – used to design LBT for co-therapy with anti-PD1